The Sai Lab published the first detailed architecture of SARS-CoV-2 on Cell
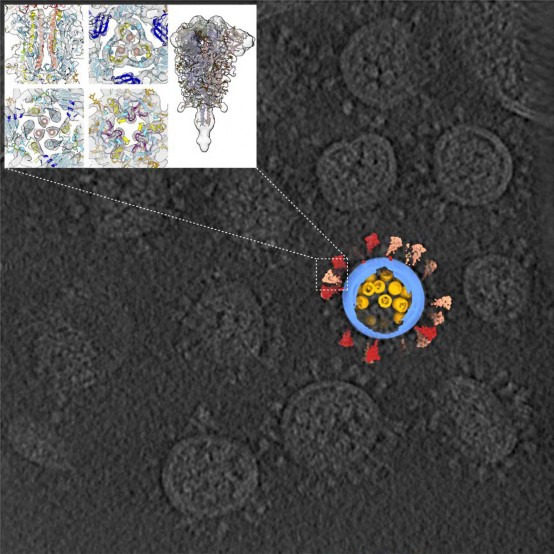
Cooperated with the Lab of Lanjuan Li, who is an Academician of Chinese Academy of Sciences, the Sai Lab published the 3D architecture of SARS-CoV-2 on the Cell magazine. This 3D result was generated with the cryo electron microscopy and subtomogram averaging.
Although it has caused 30 million cases of infection and millions of fatalities, the culprit —the SARS-CoV-2 virus remained unseen. During the pandemic, it is desired to have the image of the SARS-CoV-2 virus to educate the public, to better the strategies for disease control, as well as to support the development of the vaccines and antibodies.
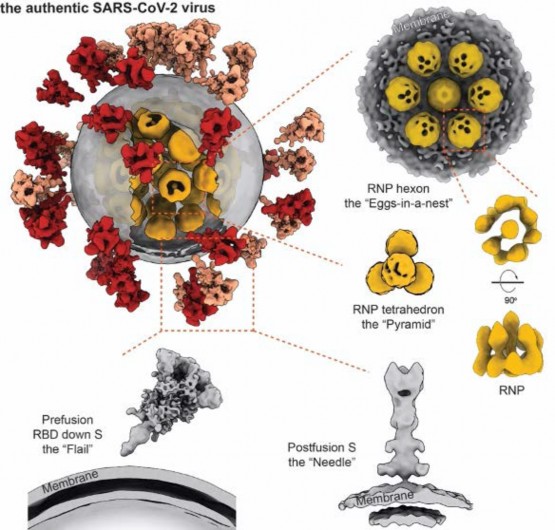
In the study, we purified and concentrated inactivated virions from Zhejiang University. Using our lab-developed, high-throughput and high-resolution cryo-ET technique, we picked out 2294 virions from 100TB data (which is the largest SARS-CoV-2 cryo-ET dataset so far) and reconstructed a representative 3D viral structure of 7.8-11 angstrom resolution. We also discovered that the SARS-CoV-2 virion has an average diameter of about 80 nanometers, about 30 spike proteins on the surface and about 30 ribonucleocapsid protein (RNP) complexes in the viral lumen. The revealed whole virion 3D image of SARS-CoV-2 showed this invisible enemy up to the public for the first time.
One of the Cell editors made high remarks that ''this work showed the most comprehensive SARS-CoV-2 image to date, and it is an extraordinary application of cryo-ET analyzing virion architecture... '' Felix A. Rey, the famous structural virologist and Academician of French Academy of Sciences, applauded by stating ''this is a milestone of the field''.
In situ spike protein structure
The coronavirus family was known because of its crown-like spike proteins on its surface. The spike protein is the key to human cells invasion through viral-cell fusion, and thus most research on anti-SARS-CoV-2 antibodies and vaccines focuses on this protein.
Our study revealed spike protein structures in three conformations, two of them being prefusion and one being postfusion. We showed that spikes adopt a random distribution on the viral surface. What's more, the spikes can rotate around their stems. This latter feature helps the ''key'' to adjust its orientation to better contact with the ''lock'', namely the receptor, so that the virus can initiate its subsequent cell invasion. However, the spike proteins are so fragile that improper inactivation or purification can cause their partial or even entire shedding and produce naked particles.
Meanwhile, one spike protein has up to 66 glycosylation modifications. These modifications can help virus evade from immune recognition. However, by publication time, no comprehensive study had described glycosylation on intact SARS-CoV-2. With the generous help from Protein Chemistry and Proteomics Facility at Tsinghua University, we reported that glycosylation on the native spike protein is slightly bulkier and more complex compared to the recombinant spike protein. But in general, this difference is minor that it should not impare the effectiveness of vaccine or antibody research based on recombinant spike proteins.
In situ ribonucleocapsid protein (RNP) complex structure
From the large sets of data, our lab picked out and analyzed around 20,000 RNPs from the viral lumen, and showed the structure and assembly of RNP complexes. These complexes locally form either a ''eggs-in-a-net'' hexon or a ''pyramid'' tetrahedron, and wind the huge viral RNA up into a secondary structure resembling a string of beads. This kind of assembly enhances the viral endurance in various environments. Furthermore, it was the first time that we visualized the interior structure of a positive-sense single stranded RNA virus.